_18.png)
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
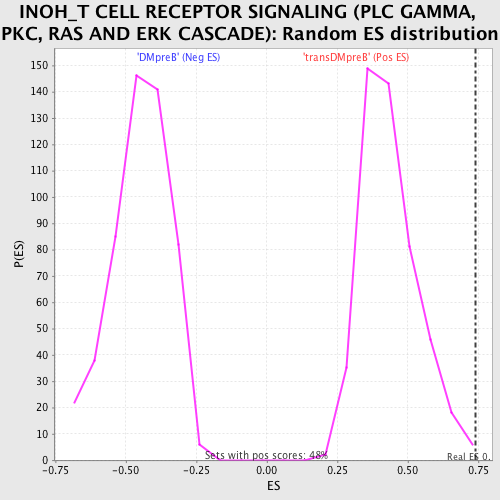
| GeneSet | INOH_T CELL RECEPTOR SIGNALING (PLC GAMMA, PKC, RAS AND ERK CASCADE) |
| Enrichment Score (ES) | 0.7401556 |
| Normalized Enrichment Score (NES) | 1.7055135 |
| Nominal p-value | 0.004166667 |
| FDR q-value | 0.077470146 |
| FWER p-Value | 0.294 |
_18.png)
| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ITPR3 | 9195 | 31 | 2.936 | 0.1118 | Yes | ||
| 2 | BTK | 24061 | 118 | 2.378 | 0.1990 | Yes | ||
| 3 | ITPR1 | 17341 | 285 | 1.959 | 0.2658 | Yes | ||
| 4 | SYK | 21636 | 513 | 1.619 | 0.3162 | Yes | ||
| 5 | TYK2 | 12058 19215 | 563 | 1.543 | 0.3732 | Yes | ||
| 6 | MAP2K1 | 19082 | 767 | 1.285 | 0.4119 | Yes | ||
| 7 | GRB2 | 20149 | 792 | 1.266 | 0.4595 | Yes | ||
| 8 | CSK | 8805 | 940 | 1.117 | 0.4948 | Yes | ||
| 9 | MAPK3 | 6458 11170 | 961 | 1.090 | 0.5358 | Yes | ||
| 10 | RAF1 | 17035 | 1186 | 0.942 | 0.5602 | Yes | ||
| 11 | JAK1 | 15827 | 1249 | 0.894 | 0.5914 | Yes | ||
| 12 | JAK3 | 9198 4936 | 1299 | 0.858 | 0.6219 | Yes | ||
| 13 | SOS2 | 21049 | 1331 | 0.845 | 0.6529 | Yes | ||
| 14 | TEC | 16514 | 1394 | 0.805 | 0.6806 | Yes | ||
| 15 | PTK2 | 22271 | 1498 | 0.741 | 0.7037 | Yes | ||
| 16 | MAP2K2 | 19933 | 1669 | 0.648 | 0.7196 | Yes | ||
| 17 | ARAF | 24367 | 1729 | 0.613 | 0.7402 | Yes | ||
| 18 | PTK2B | 21776 | 2197 | 0.385 | 0.7299 | No | ||
| 19 | RASGRP1 | 5360 14476 9694 | 2883 | 0.220 | 0.7015 | No | ||
| 20 | ZAP70 | 14271 4042 | 2886 | 0.219 | 0.7099 | No | ||
| 21 | JAK2 | 23893 9197 3706 | 3607 | 0.130 | 0.6762 | No | ||
| 22 | LAT | 17643 | 3622 | 0.129 | 0.6804 | No | ||
| 23 | PLCG1 | 14753 | 5539 | 0.045 | 0.5791 | No | ||
| 24 | SOS1 | 5476 | 5727 | 0.041 | 0.5706 | No | ||
| 25 | ITK | 4934 | 8021 | 0.007 | 0.4475 | No | ||
| 26 | SRC | 5507 | 10688 | -0.026 | 0.3051 | No | ||
| 27 | CD4 | 16999 | 12204 | -0.054 | 0.2257 | No | ||
| 28 | NRAS | 5191 | 12495 | -0.061 | 0.2124 | No | ||
| 29 | KRAS | 9247 | 15861 | -0.392 | 0.0465 | No | ||
| 30 | FYN | 3375 3395 20052 | 15886 | -0.398 | 0.0606 | No | ||
| 31 | HRAS | 4868 | 17006 | -0.832 | 0.0325 | No | ||
| 32 | LCK | 15746 | 17991 | -1.398 | 0.0336 | No |
_19.png)